Research
Protein Ultrastructure by Cryo Electron Tomography
The Metskas lab is interested in how proteins assemble into ultrastructures in order to perform specific functions. These ultrastructures may be as simple as enzyme polymerization, or as complex as an entire virus, but they combine the intrinsic properties and motions of their subunits in order to perform a complex function that a single unit would not be capable of.
We use cryo electron tomography to visualize ultrastructures in their native environment, and subtomogram averaging to build higher-resolution maps of the subunits. We then use the information on the protein positions and orientations to build biophysical models of the functioning ultrastructure.
Cryo Fluorescence Microscopy
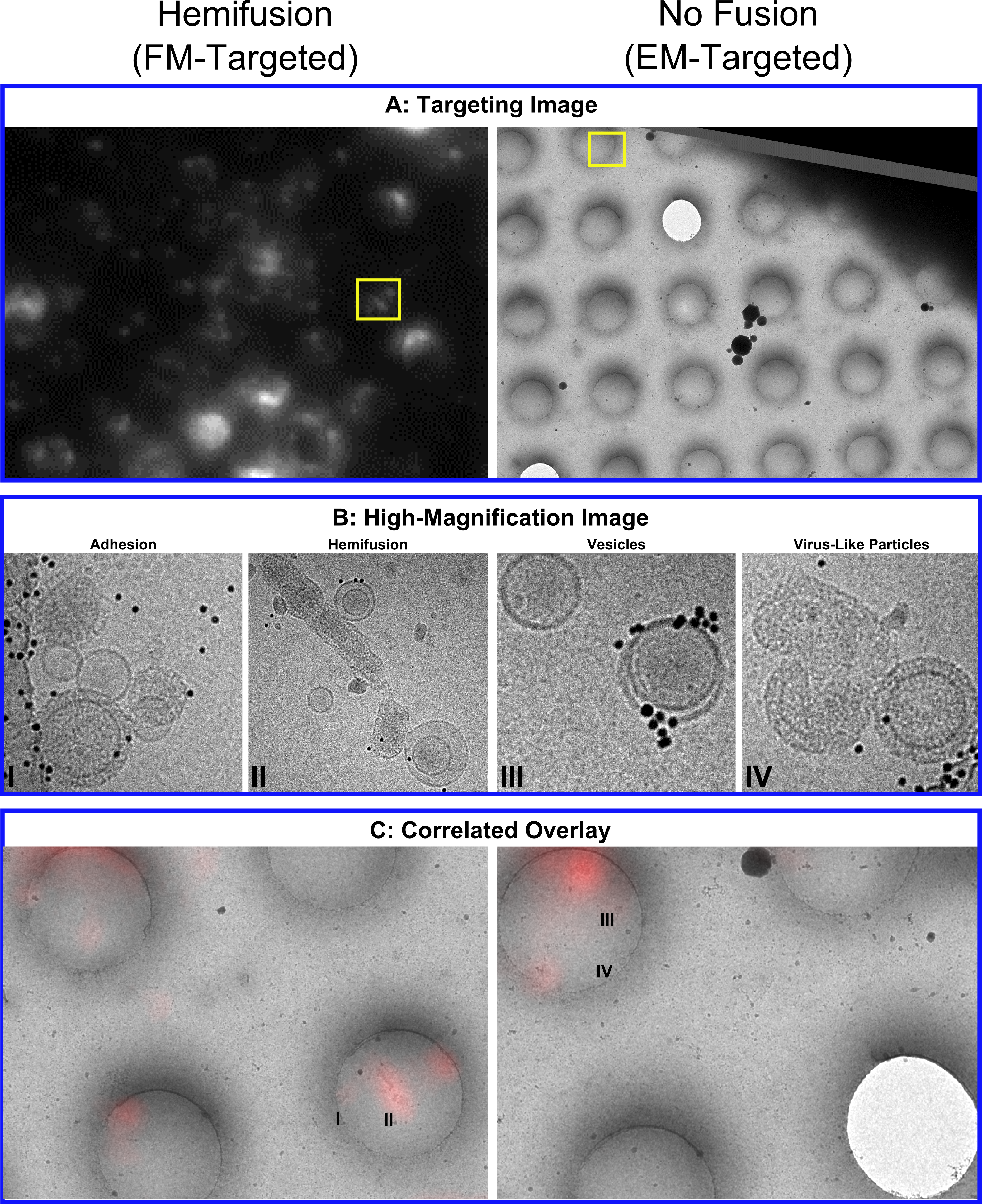
In order to identify and image rare events on a cryo EM grid, we correlate the cryo electron microscopy with a cryo fluorescence image of the same grid (cryo CLEM). This allows us to combine the resolution of tomography with the specificity of fluorescence. The use of fluorescence can serve as a link between cryo EM and room-temperature fluorescence-based studies, by linking the fluorescence signal with the micrographs.
Temperature affects fluorophores in unique ways, many of which are poorly understood. Our group members are developing new applications that work under cryo conditions, which we adapt as tools for use in cryo-fluorescence microscopy.